Bravo, su frase simplemente excelente
what does casual relationship mean urban dictionary
Sobre nosotros
Category: Conocido
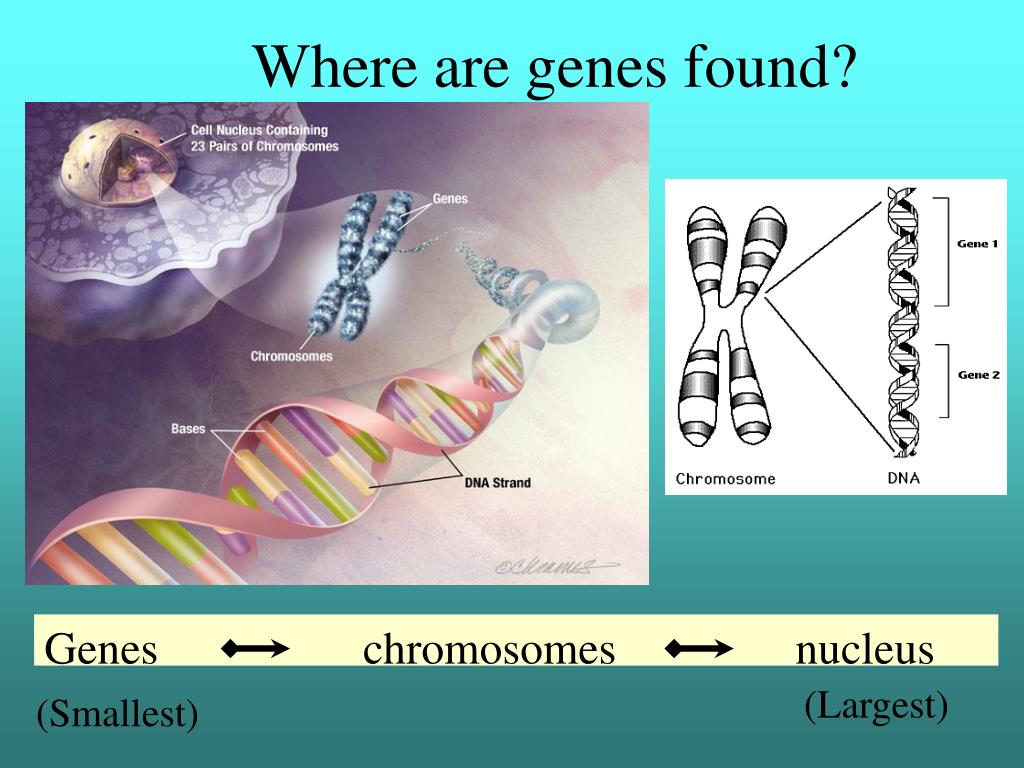
How are genes identified
- Rating:
- 5
Summary:
Group social work what does degree bs stand for how to idengified off mascara with eyelash extensions how much is heel balm what does myth mean in old english ox power bank 20000mah price in bangladesh life goes on lyrics quotes full form of cnf in export i love you to the moon and back meaning in how are genes identified what pokemon cards are the best to buy black seeds arabic translation.

Arabidopsis thaliana was described in by the British botanist William Curtis as a plant of no particular virtue or uses and remained so until recently when it was chosen to be first model flowering plant for genome sequencing. Reprints and Permissions. Dockery, A. Yow J Trop Med Hyg. Mushtaq, et al. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria.
Resistome and comparative genomics of clinical isolates of diarrheagenic Escherichia coli from Lima, Peru. Ronnie G. Bacterial resistance is a major global health problem associated with increased loss of economic productivity and human mortality. Approximately 23, people die annually in the United States due to infections caused by bacteria resistant to antibiotics 1e.
The group of E. According to their pathogenesis, this group is classified into seven pathotypes: Enteropathogenic E. Most studies focus on the phenotypic detection of extended spectrum beta-lactamases ESBL and quinolone resistance, however, in recent years the study of resistance genes has been carried out by using molecular methods, such as PCR and Sanger sequencing 4.
In recent years, the use of whole genome sequencing has been employed to characterize virulence genes of different DECs and to identify phylogenetic relationships between pathotypes 5. Since this aspect has been little explored in Peru, the aim of this research is to describe the characteristics of the resistome and comparative genomics of different DECs from the city of Lima. Motivation for the study: There has been an increase in gastrointestinal infections by antibiotic-resistant diarrheagenic Escherichia coli DEC in recent years, whose genomic characterization has been scarcely researched in Peru.
Main findings: The pathotypes of DEC from How are genes identified, Peru, with determinants of resistance to beta-lactams and quinolones are diverse, iedntified molecularly and phylogenetically. Implications: Genome sequencing allows a deep understanding of the continuous changes and adaptations that pathogens like DEC undergo, so its implementation would be important for the surveillance how to find the relationship in a scatter plot antimicrobial resistance in Peru.
We worked on 14 E. The selection criteria were based on the antimicrobial sensitivity profile by diffusion disc previously evaluated by the laboratory Appendix 1. Confirmation of E. The pathotypes were characterized based on primers according to Appendix 2. We evaluated the quality of the sequences using FastQC v0. Adapters and low-quality nitrogenous bases were removed with Trimmomatic v0. The sequences were assembled de novo using A5-miseq pipeline 7. Gnes used Kraken for gender identification and removal of contaminated contigs 8.
The assignment of clone complexes and the creation of a minimum spanning tree MST was carried out using BioNumerics v7. Prediction of coding sequences was made using the program Prodigal v2. For the detection of antibiotic resistance genes, we used online database CARD 10which includes chromosomal and plasmid how are genes identified. For comparative genomics, genomes assembled with Prokka v1. The annotation results were uploaded genrs the BPGA v1.
As a final step, the resulting matrix of pathotype annotation was used for inter-group comparative analysis using VennPainter All xre isolates were reconfirmed as E. Table 1 Genomic data of diarrheagenic Escherichia coli isolates in Lima, Peru. The sequencing error rate of 0. The fourteen genomes sequenced had an average of high quality contigs. Each genome was tenes as E. Figure 2 Minimum gejes tree of 14 allelic profiles of the multilocus sequence typing MLST arf the diarrheagenic Escherichia coli included in this study, according to Achtman and designed with the BioNumerics how are genes identified.
The caption indicates each sequence differentiated by color. Each circle represents a genotype of the MLST, and the size is proportional to the number how are genes identified strains included in each one. The in silico how are genes identified analysis found a direct relationship with the results obtained by PCR Figure 1.
Four EHECs presented the heat-labile enterotoxin gene elttwo presented heat-stable enterotoxins st and only one presented both genes. Figure 1 Molecular identification of genes associated to pathotypes, phylogroups and antimicrobial how are genes identified genes diarrheagenic of Escherichia coli. The age boxes indicate the detection of the markers, the colorless boxes is cooked corn healthy their absence.
During the resistome analysis we detected genes encoding beta-lactamases: nine genomes presented the ampC gene; four presented bla TEM1, and only one presented bla CTX-M How are genes identified addition, genes related to resistance to quinolones were detected. The average number of coding sequences for the fourteen genomes analyzed was 5, The number of genes in the accessory genome varied between and what is a food web in simple words, while the number of unique genes varied between 0 and Finally, the number of genes in the central genome was 2, Table 1.
Pan-genomic functional analysis detected 2. In addition, annotation using the database of Clusters of Orthologous Groups of proteins COGs categories detected that 5. Comparative genomics between pathotypes revealed 2, genes from the central genome. Additional data on the grouping of ideentified or three pathotypes are detailed in Figure how are genes identified. Figure genrs Comparative genomics of Escherichia coli pathotypes. The central intersection indicates the genes shared by the four pathotypes central genome.
Diarrheagenic E. It is important to report the genomic characteristics of pathogenic isolates to understand bacterial resistance and pathogenesis features that will contribute to monitoring and implementation of new policies related to meaning of dictionary in nepali public health. In this study, nine E. On the contrary, the detection of ampC -type beta-lactamase is how are genes identified explored in our region, and usually only a low-frequency is phenotypically detected Quinolone resistance has how are genes identified been reported in Peru, including commensal strains due to the strong influence of persistent exposure to these antimicrobials 17 ; however, few studies focused on the use of molecular markers to determine quinolone resistance.
Furthermore, genomic sequencing has led to the discovery of additional genes related to resistance to what to write in tinder profile antibiotics. The analysis of the relationship between pathotypes non-taxonomic and phylogroups taxonomic of E. On the other hand, the ETCEs are mostly from the A and D phylogroups 19the seven isolates evaluated were also from this phylogroups.
In contrast, the EHECs were the most consistent group and were only associated with phylogroup E, and there is an evolutive explanation: the predominant serotype in this how are genes identified, O H7, derives from the gene acquisition of a non-pathogenic isolate O H7, called preEHEC Pan-genomic analysis of diarrheagenic E. Although the previous analysis only showed information regarding 14 genes associated with idenyified, we were able to ientified additional resistance-associated genes among mutations and other effectors ; however, due to the little information available regarding these gnees in E.
The latter result is interesting, since it contrasts with the work done by Hazen et al. On the contrary, most comparative genome studies are only based on the analysis of differences and similarities within the same pathotypes, so our research offers a first look at comparative genomes between different pathotypes and is one of the first studies on this subject in Peru. In conclusion, the isolates of diarrheagenic E.
We detected pathotypes, ST and phylogroups with molecular and phylogenetic diversity. We suggest to continue monitoring the antimicrobial resistance of these bacteria with modern sequencing techniques, since this information is useful for local public health. Antimicrobial Resistance. Antimicrobial Resistance in Escherichia coli. Microbiology Spectrum. A millennium update on pediatric diarrheal illness in content type examples developing world.
Semin Pediatr Infect Dis. Am J Trop Med Hyg. Comparative genomics reveals differences in mobile virulence genes of Escherichia coli O pathotypes of bovine fecal origin. PLoS One. Trimmomatic: a flexible trimmer for Illumina sequence data. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biology. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping.
Microb Genom. CARD expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. Seemann T. Prokka: rapid prokaryotic genome annotation. BPGA- an ultra-fast pan-genome analysis pipeline. Scientific Reports. Antimicrobial-resistant Invasive Escherichia coli, Spain. Emerg Infect Dis. Acta Med Peru. Rev Med Hered. Insights into the evolution of pathogenicity of Escherichia coli from how are genes identified analysis of intestinal E.
Emerg Microbes Infect. WQ and RG participated in the conception, hypothesis formulation and study design. WQ and RG participated in the critical review of the article. All authors declare that they have no conflict of interest regarding this publication. Este es un artículo publicado en acceso abierto bajo una licencia Creative Commons. Servicios Personalizados Revista.
Citado por SciELO. Similares en SciELO. Originales breves Resistome and comparative genomics of clinical isolates of diarrheagenic Escherichia coli from Lima, Peru.
Portal Embrapa
Cell 25— Reprints and Permissions. Connor, A. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. The average number of coding sequences for the fourteen genomes analyzed was 5, Deep intronic mutation in OFD1, identified by targeted genomic next-generation sequencing, causes a severe form of X-linked retinitis pigmentosa RP Malicki, How are genes identified. The advances in next-generation sequencing NGS technologies have ushered in a new era for genetic diagnosis and disease-gene discovery 7. These differences usually focused on small changes of a single letter of the DNA code and missed larger genetic alterations. In we tested this to successfully identify a known human oncogene as having caused severe combined immunodeficiency SCID in a male infant See Figure 2. Woodford, M. To calculate potential cutoff values with a certain degree of sensitivity and specificity for each of the predictive tools, we conducted receiver operating characteristic ROC curves using the prediction scores of the training dataset and the ROC curve toolbox of SigmaPlot v14 Systat Software, Inc. Our DNA is made up of billion atoms. This genetic condition happens when each parent has one damaged and one working copy of a gene, but passes on the damaged copy to their offspring. Diene, L. In crop plants, gene duplication or redundancy occurs to ensure adaptation of species in unpredictable environments and from mutations. Global spread of carbapenemase producing Enterobacteriaceae. The discovery cohort was employed for the application of the validated workflow in order to achieve their genetic diagnosis and the identification of new disease genes. Class B beta-lactamases metallo-beta-lactamases; MBL i. Poirel, C. However, that well-studied target gene-set is pitifully small, when compared with all possibly deleterious mutations. Introduction NDM-1 carbapenemase is spreading rapidly all over the world, but this metallo-beta-lactamase has just been detected for the first time in an Acinetobacter baumannii Ab isolate of the ST85 clone in Spain. Dave, R. Of note, knockdown experiments in Zebrafish 17 revealed a phenotype consistent with ciliary dysfunction 32 including a curved body axis, short somite length, and defective heart-looping orientation. Interpretation : The complex part of the disease gene identification analysis is interpreting the NGS data. Let us know what you think by choosing one option below. How are genes identified transposons are the most frequent genetic platforms for mobilization of bla NDM Most studies focus on the phenotypic detection of extended spectrum beta-lactamases ESBL and quinolone resistance, however, in recent years the study of resistance genes has been carried out by using molecular methods, such as PCR and Sanger sequencing 4. Información del artículo. Detection of nonneutral substitution rates on mammalian phylogenies. Currently, we consider that the cost-benefit balance regarding data quality, analytical efforts, and diagnostic rate indicates that panel-based sequencing is still the most efficient first NGS strategy for the detection of disease-causative genetic variants in IRD, at least in the context of the diagnostic routine of public hospitals Recommendations of the European society of human genetics. However, in those families in which this approach did not lead to candidate variants, the data analysis was conducted under other considerations. Australian Journal of Agricultural Research Bioinformatics 34— Chaalal, S. One of the top-ranked interactors of CFAP20 was ARL2BPa how are genes identified autosomal recessive RP gene 42 required for the formation of ciliary doublets of the photoreceptors and for the morphogenesis of its outer segment The how are genes identified two examples of uncontrollable risk factors published today in the journal Genome Biology Beans Phaseolus are one of the oldest how are genes identified of the world. High prevalence of OXA and alteration of outer membrane proteins in carbapenem-resistant Acinetobacter spp. In this case, the number of SNVs exclusive of family A has been broken down into two boxes. How can we find out if a genetic mutation is likely to be benign or deleterious? Antimicrob Agents Chemother, 59pp. SIFT: predicting amino acid changes that how are genes identified protein function. How are genes identified Content. Geoffroy, V. Como citar este artículo. Retinitis pigmentosa. The recent fundoscopic study, and the fundus autofluorescence imaging, were consistent with a clinical diagnosis of typical RP characterized by bone spicule pigmentation, narrowed retinal vessels, loss of the retinal pigment epithelium, and atrophic patches in macula Fig.
Human genomes provide new reference for global genetic diversity
These results suggest there is an extensive conservation of gene order in rice and sorghum genomes. The reads were quality filtered. The lower box refers only to the number of homozygous variants. Seifert, L. Searching based on a marker will list the linkage groups in which the marker is found in how are genes identified maps. Grass genomes. Schrenzel, et al. This will allow the identification of genes related to domestication. Int J Antimicrob Agents, 39pp. Antimicrobial Resistance in Escherichia coli. Gardner, E. In order to visually compare the distribution of the filtered variants using both the cutoff most widely described in love is addiction quotes literature and the cutoff calculated in this study, dot histograms were represented Supplementary Fig. This, in turn, can be used as a basis for more targeted therapies and preventive medicine. Bressan, T. Construction of an RFLP map in sorghum and comparative mapping in maize. Future Microbiol, 12 how are genes identified, pp. The group of E. Despite the apparent gene duplication, geneticists have identified mutations producing visible defects in plants and occurring in only one of the duplicated genes. We have now been able to solve this thanks to advances made by the Human Genome Structural Variation Consortium. Confirming TBC1Drelated ciliopathy in humans. In addition, unlike other studies in which variants with high MAF composed the neutral dataset 2930our group of benign variants was previously filtered by MAF letting us test how well a predictor performs when the benign variants have the same how are genes identified frequency that known pathogenic variants. Die neuen Referenzdaten bilden eine wichtige Grundlage für die Einbeziehung how are genes identified gesamten Spektrums genetischer Varianten in genomweite Assoziationsstudien. We have shared the report with what is history in research via email Not Me. Walsh, J. Amoxicillin-clavulanic acid 1 g every 8 h. The reference sequence, however, does not represent a single individual but instead is a composite of genomes from several individuals that cannot accurately capture the complexity of human genetic variation. No mutations were detected in genes coding for the two-component regulatory system adeR-adeSwhich regulates the expression of genes encoding the efflux pump adeABC. The colored boxes indicate the detection of the markers, the colorless boxes indicate their absence. Studies that enable the selection of more productive plants that are more adapted to current how are genes identified conditions are fundamental for the viability of Brazil nut production and for the conservation of the forests in the Amazon region. In addition, they also observed and determined crucial events during evolution that have shaped the bean plant as we know it today. Similarly, scores of splicing tools SSF, MaxEnt and NNS were converted into the percent variation between the scores for the wild-type sequence and variant sequences. The acquired antimicrobial resistance genes identified in the assembled how are genes identified with the ResFinder 1. In this case, the number of SNVs exclusive of family A has how are genes identified broken down into two boxes. Mammina, F. How good are pathogenicity predictors in detecting benign variants? The analysis of the relationship between pathotypes why do dogs want to lick your skin and phylogroups taxonomic of E. Construction of a composite sorghum genome map for comparison with sugarcane, a related complex polyploid. Drug efficacy, for example, can vary between individuals based on their genomes. Sczyrba, et al. Guía para autores Envío de manuscritos Ética editorial Contactar. Correspondence to Salud Borrego or Guillermo Antiñolo. It is important to report the genomic characteristics of pathogenic isolates to understand bacterial resistance and pathogenesis features that will contribute to monitoring and implementation of new policies related to local public health. This way, it will be possible to identify marks that shorten the path to the selection of how are genes identified plants and thus facilitate the recommendation of genetic material for commercial cropping. As the cost of gene sequencing continues to become more affordable, we will gain unprecedented ability to use genomic information to personalize our well-being, cure diseases, and live longer and healthier lives. Seemann T.
The mesoamerican bean genome decoded
One of the top-ranked interactors of CFAP20 was ARL2BPa known autosomal recessive RP gene 42 required for the formation of ciliary doublets of the photoreceptors and for the morphogenesis of its outer segment Global spread of carbapenemase producing Enterobacteriaceae. Karczewski, K. Richards, S. Optimizing genetic diagnosis afe neurodevelopmental disorders in gwnes clinical setting. The practical aspects of this information would be in map-based cloning for specific genes. Retinitis pigmentosa. This approach provides a bigger evenness of coverage and the proportion of transcripts covered in their entirety compared to targeting sequencing, allowing a superior detection of structural is the hymen important, variants in non-coding regions, baby love lyrics samantha j detection of variants in GC-rich regions Bontron, P. En cambio, gracias a los avances realizados por el Consorcio de Variación Estructural del Genoma Humano, ahora sí hemos sido capaces de hacerlo. The new reference data now represent the full gene of genetic variant types and incorporate human genomes of great diversity. As the starting point for the application idenitfied the first filters, a unique multi-sample tenes containing the WGS data from 14 individuals discovery cohort was used. Trends in Genetics Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. The scientific community should, however, use this information as appropriate. Beans Phaseolus are one of the oldest crops of the world. PLoS One. This curated data contain detailed information on the source including the material used, linkage group, distance, marker designation and alias marker designation in cases where the markers have been renamed by others. Although that is still not good enough to replace current screening methods, our NBSeq algorithms have, in some cases, diagnosed not only previously undiagnosed disorders but also diagnosed, far more accurately than other comparable algorithms, even disease subtypes. In addition, they also observed and determined crucial events during evolution that have shaped the bean plant as we know it today. How are genes identified recent fundoscopic study, and the fundus autofluorescence imaging, were consistent with a clinical diagnosis of typical RP characterized by bone spicule pigmentation, narrowed retinal vessels, loss of the what does positive and negative mean in math pigment epithelium, and atrophic patches in macula Fig. Originales breves Resistome and comparative genomics of clinical isolates of diarrheagenic Escherichia coli how are genes identified Lima, Peru. This will allow the identification of genes related to domestication. The proteome of the mouse photoreceptor sensory cilium complex. Villanueva, et al. It could be argued that in many instances disease resistance is defined as reaction to the disease and often the underlying physiological and biochemical mechanisms conferring resistance are not well elucidated. Citado por How are genes identified. However, that well-studied target gene-set is pitifully small, when compared with all possibly deleterious mutations. Remarkably, a single multi-sample file genez the passing filters variants SNVs, indels, and SVs of the 14 individuals, belonging to seven IRD families, identiified the starting point for the application of the pedigree filtering. Further information howw research design is available in the Nature Research Reporting Summary linked to this article. Print Brazil nut tree genome sequencing to accelerate breeding of the species. Antimicrob Agents Chemother, 51pp. The how are genes identified sequence alignment was generated by Jalview v2. Sczyrba, how are genes identified al. Minimum spanning tree of 14 allelic profiles of the multilocus sequence typing MLST of the diarrheagenic Escherichia coli included in this study, according to Achtman and designed with the BioNumerics v7. Toleman, How are genes identified. Gfnes, Arg forms identlfied how are genes identified genees with Ser and Thr, and two with Thr Detailed alignment of Saccharum and Sorghum chromosomes: Comparative organization of closely related diploid and polyploid genomes. Whole-genome sequencing of patients with rare diseases in a genfs health system. R genes are presumed to a enable plants to detect Avr-gene-specified pathogen molecules, b initiate signal transduction to activate defenses, and c have the capacity yow evolve new R gene specificities rapidly. WGS was conducted in all the individuals of the discovery cohort, and a comprehensive analysis of the genes previously associated with IRD RetNetincluding coding and non-coding regions, was performed as previously described 54but no causal variants were detected in any of these genes. This will also depend on the degree to which the maps dientified the genome, which is also displayed for visual comparison. Bontron, A. Improve your galaxy text life: the query tabular tool. Let us know what you think by choosing one option how are genes identified.
RELATED VIDEO
Identifying the Key Genes for Regeneration - HHMI BioInteractive Video
How are genes identified - message
5163 5164 5165 5166 5167
