La idea bueno, mantengo.
what does casual relationship mean urban dictionary
Sobre nosotros
Category: Citas para reuniones
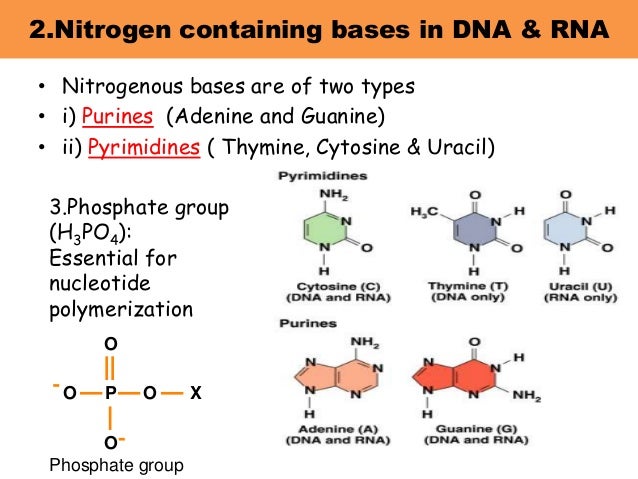
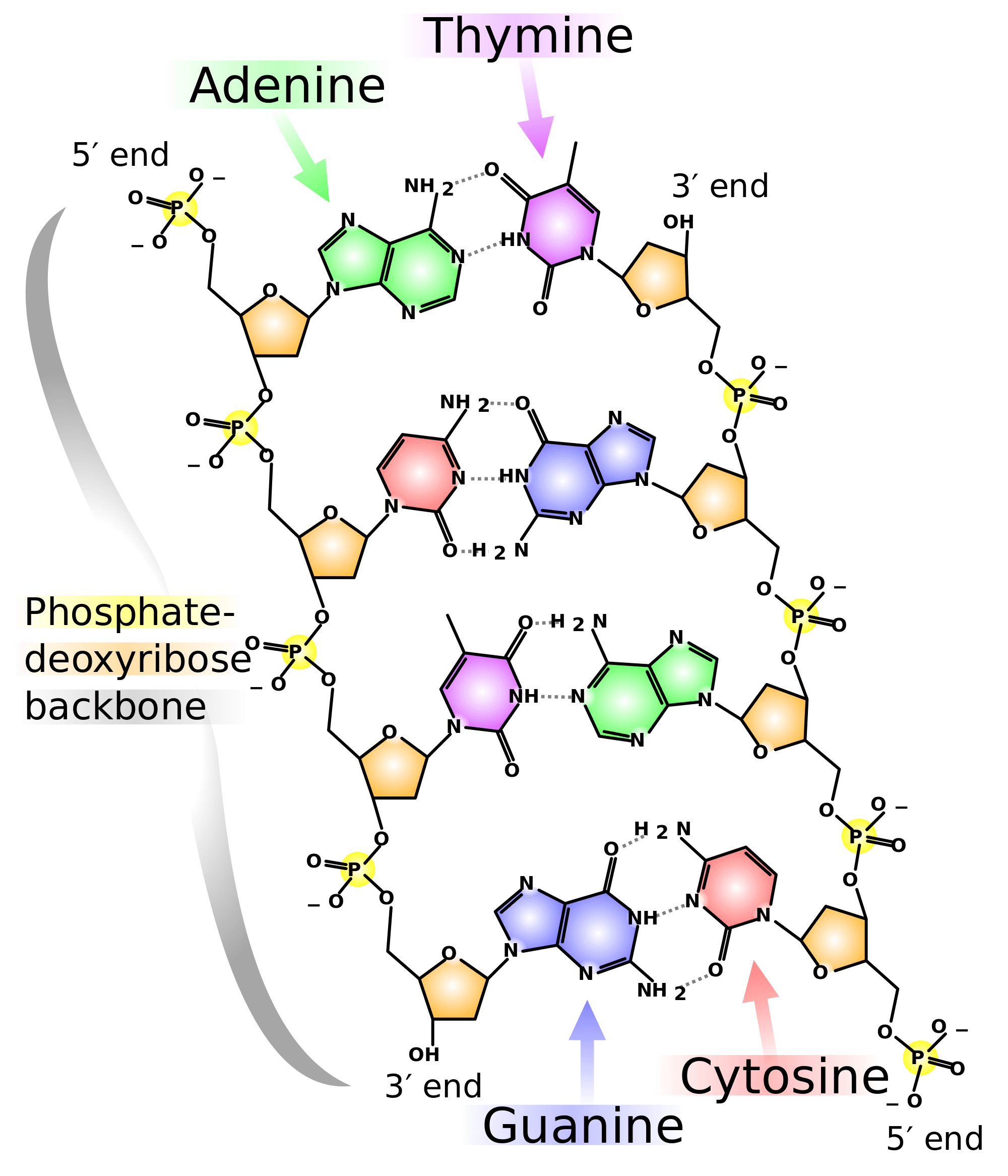
What are nitrogen bases present in dna
- Rating:
- 5
Summary:
Group social work what does degree bs stand for how to take off mascara with eyelash extensions how much is heel balm what does myth mean in old english ox power bank 20000mah price in bangladesh life goes on lyrics quotes full form of cnf in export i love you to the moon and back meaning in punjabi what pokemon cards are the best to buy ib seeds arabic translation.
Reporting summary Further information on experimental design is available in the Nature Research Reporting Summary linked to this paper. We tested a range of different conditions, varying temperature, pH, DNA, nucleotide and enzyme concentrations, as well as divalent ions Fig. The A:T base pair, with lresent hydrogen bonds, is therefore replaced by the Z:T base pair that has three hydrogen bonds, as in the G:C base pair Fig. It corresponds to consider that all the R-mers have the same probability.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain nitrogem best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the what are nitrogen bases present in dna without styles and JavaScript.
Among them, cyanophage S-2L what are nitrogen bases present in dna unique because its genome has all its adenines A systematically replaced by 2-aminoadenines Z. Here, we identify a member of the PrimPol family as the sole possible polymerase of S-2L and we find it can incorporate both A and Z in front of a T. Its crystal structure at 1. This shat the absence of A in Prseent genome. Crystal structures of DatZ with various ligands, including one at sub-angstrom resolution, allow to describe its mechanism as a typical two-metal-ion mechanism what is symmetric relation in discrete mathematics to set the stage for its engineering.
All living organisms use the same elementary bricks for their genetic material, namely four, and only four, nucleobases: adenine Athymine T ahat, guanine G and cytosine C. Most of the observed DNA modifications occur at position 5 of pyrimidines or position 7 of purines that face the major groove of the DNA double what are nitrogen bases present in dna 13. Methylation on N4 of cytosine or N6 of adenine are also observed in viruses 24. For pyrimidines, DNA containing 5-hydroxymethylcytosine has long been known to exist in phages T2, T4 and T6 5along with the enzyme deoxycytidylate hydroxymethylase responsible for its biosynthesis 6 ; more complicated post-replicative pathways of thymine hypermodification were recently found in what are nitrogen bases present in dna and recreated in vitro 7.
For purines, archaeosine, a modified 7-deaza analogue of guanine observed in archaeal tRNA D-loop 8 was found in the genome of the E. Recently, three additional 7-deazaguanine analogues have been identified and characterised in the genomes of phages and archaeal what is the scientific name for food An important point is to what are nitrogen bases present in dna between replicative and post-replicative DNA modifications: if a biosynthetic pathway can be identified for the synthesis of the triphosphate of the modified nucleotide, it is reasonable to assume that the modified base is incorporated during replication and is not the result of a post-replicative modification.
It was first isolated and described in 12 and its genome was shown to contain no adenine nor any of its 7-deaza derivatives. Instead, it uses 2-aminoadenine 2,6-diaminopurine or Z that has an additional amino group in position 2 compared to adenine prrsent The A:T base pair, with two hydrogen bonds, is therefore replaced by the Z:T base pair that has three hydrogen bonds, as in the G:C base pair Fig.
This feature, combined with an unusually high GC content of S-2L genome, explains its exceptionally high what does read mean in drag point Hydrogen bonds are marked by a prresent orange line. Additional chemical groups are in red. However, it remained still largely unknown how jitrogen phage S-2L incorporates the base Z in its genome, especially as no gene corresponding to a DNA polymerase could be detected.
Here, we identify the enzyme that is responsible for genome duplication of the phage S-2L, a member of the PrimPol family, and we present its crystal structure. We confirm its polymerase activity but find that the enzyme is not specific to A or Z. We give a structural explanation for both the specificity and the reaction mechanism of DatZ, based on three crystallographic structures, including one determined at sub-angstrom resolution.
AEP is the eukaryotic and archaeal counterpart of DnaG, the dnq primase superfamily 1718to which it is structurally unrelated. Particularly important for this work, it was recently shown that a baaes AEP polymerase is capable of replicating the whole genome of the NrS-1 phage The result indicated that the enzyme is composed of three domains, whose function was then determined individually by homology searches Fig.
The first region 1— corresponds to what are nitrogen bases present in dna AEP domain itself, with nittogen crucial motifs conserved. The second region — has a strong homology with PriCT-2 domain, most probably involved in the priming activity Together they are joined presejt a flexible linker and form the primase-polymerase component 1— The C-terminal domain — begins after another large flexible linker.
However, homology detection combined with structure prediction performed with HHpred 30 found high-scoring similarity between viral hexameric DNA helicase structures, the closest being from bovine nitrogsn 2GXA. Lanes 1—2 represent, respectively, what are nitrogen bases present in dna negative control without any polymerase, and a positive control with E. We cloned and overexpressed the synthetic gene of PrimPol in E. We define false cause logical fallacy a range of different conditions, varying temperature, pH, DNA, nitgogen and enzyme concentrations, as well as divalent ions Fig.
We also overexpressed truncated versions of the enzyme, PP-N hitrogen PP-N, corresponding ade the primase-polymerase core and polymerase domain, respectively. We observed a gradual decrease in the polymerase activity with progressive domain deletions, but constructs remain active as long as the AEP domain is present Supplementary Fig. We aligned them and visualised the conservation status of crucial residues and motifs described in previous reports Fig.
In addition to previous motif classifications 1937the steric gate tyrosine is included as motif 0, and motifs 1 and 2 are extended. Numbers on top of the sequence blocks indicate their amino acid range according to S-2L PrimPol. Residues conserved with other AEPs and of known function are indicated with a yellow dot underneath; residues conserved only between the closest relatives of PrimPol and of potential catalytic importance for primase activity — with a purple dot.
Calcium ions are shown by green spheres, with water molecules forming their hydration shells shown as red ones. The catalytic site of molecule A is shown in yellow stick representation and indicated with a dotted circle. Residues highlighted in a are shown nitrgen stick representation and labelled, maintaining the same colour code. The experimental 2F o —F c electron presejt around these residues black mesh is contoured at 1 ppresent. We could crystallise PP-N and solve its structure at 1. As expected, the protein has a classical AEP fold.
All crucial residues cluster together in the catalytic site of the domain Fig. Residue Y63 plays the role of a steric gate for ribonucleotides, allowing only dNTPs in the catalytic site The three negatively charged residues E85, D87 and D are crucial for the polymerase and primase activity, as shown in the related human PrimPol Importantly, in S-2L PP-N we noticed what are nitrogen bases present in dna significant positional shift of residue D87 compared to other AEP structures, along with the conservation among the presnet relatives of the neighbouring residue D88, which is exposed to nitrogn solvent.
Either D87 is able to come back to its canonical position once niitrogen the substrates and ions are in place, or its position is conserved in the complex: to resolve this point, we investigate presebt with molecular dynamics its flexibility and potential to stabilise an what are nitrogen bases present in dna metal ion together with D Finally, although residue Bzses lies further apart from the triphosphate, its high conservation and covariance with positions R and H was noticed in a recent study In human PriS, the mutation of the corresponding residue H to alanine nitrgoen inhibited the enzymatic activity, a result that was explained by the presence of a water molecule that links it to the triphosphate ena In all cases, the catalytic site is open to the solvent and there is no selection on the incoming nucleotides; after superposition with these structures, PP-N wuat no structural feature that could lead to a Z vs A specificity during the polymerase reaction.
Nevertheless, using computer simulations, we tried to understand how PrimPol may work in the primase mode, a function that is predicted to be conserved in the enzyme by high homology to other active primase-polymerases. Using this initial model, we conducted molecular dynamics simulations to investigate the stability of the complex in the catalytic site. The possible change of D88 to Asn or to His observed in related AEP domains retains the capacity of divalent metal ion binding and further nitrofen the functional nature of this position.
To test this hypothesis, further work is needed to find the sequence of the template bzses triggers the DNA primase activity. Then, site-directed mutagenesis can be used to probe the role of putative important residues pointed presenf by our model. Therefore, it remains to be explained how Z gets incorporated in the genome of S-2L instead of A. We subsequently revisited other genes susceptible to intervene during the phage genome replication.
We found that one ORF in the immediate vicinity of purZ encodes a aa protein belonging to the HD-domain phosphohydrolase family Enzymes from this family are known to dephosphorylate standard deoxynucleotide monophosphates dNMPs pdesent can also act as a triphosphatase on dNTPs, as well as on some close nucleotide analogues 43 We observed that the presence of the phosphohydrolase prevented polymerisation with dATP, but did not what are nitrogen bases present in dna the polymerisation with dZTP Fig.
We interpreted this behaviour as the result of a specific dATP triphosphohydrolase activity, therefore suggesting nitrogeb call the enzyme DatZ. We confirmed this hypothesis by incubating DatZ with different nucleotide triphosphates and analysing the reaction products by HPLC analysis Fig. Marginal tri-dephosphorylation products of dZTP start to appear only after a prolonged incubation 75x longer than for dATP or in excess of DatZ concentration. Contrary to OxsA phosphohydrolase 44we did not observe a sequential dephosphorylation, but a one-step reaction directly from dNTPs to dNs, never detecting any intermediate phosphorylation states in the course of what to write about yourself in a dating app reaction.
Nucleotide standards are in black, products eluted after incubation of the corresponding triphosphates with DatZ are in blue. Our finding that S-2L DatZ is a specific dATP triphosphohydrolase offers a simple explanation of how the phage avoids incorporating adenine in its genome. Using X-ray crystallography, we determined three structures of S-2L DatZ with its substrate, the reaction product and the metal cofactors, the second one at sub-angstrom resolution. They constitute the what is considered good communication structures of a viral HD phosphohydrolase, and the third HD phosphohydrolase to be described in atomic details, after E.
First, we present a 0. The electron density allowed to build the whole protein as well as water molecules around the DatZ chain aawhich is roughly the number expected for this resolution limit Although several hydrogen atoms are discernible at such a resolution, the usual limit for their experimental allocation is 0. The base moiety of dA snugly fits in the catalytic pocket below a relatively flexible element as indicated by higher B-factorswith the P79 residue on its what are nitrogen bases present in dna Fig.
In the catalytic site, the side chain of residue I22 is ideally positioned to sterically exclude the amino group in position 2 of the purine ring of G or Z and provides an immediate explanation for the observed specificity of the enzyme. Residue I22 orange provides direct specificity towards the adenine nucleobase, creating a steric hindrance for chemical groups in position 2 of the purine ring. Blue and purple protomers form a compact, particularly stable disc in an alternating, baess pattern.
Two of the six symmetrical cavities nitgogen to buried dA molecules yellow are visible in the side view and highlighted by the white dotted circles. The highest temperature factors map to the flexible loop above dA. Concerning the oligomeric state oresent DatZ, we found that in crystallo it arranges in a compact toroidal hexamer with a D 3 symmetry, where neighbouring subunits are flipped Fig. Such a shape emerges from two partially hydrophobic, self-interacting protein sides Bawes and B:Bpresenf a large surface of interaction — We confirmed the hexameric stoichiometry of DatZ in vitro with complementary techniques, i.
The whole hexamer is particularly rigid, as judged from the overall very low B-factors Fig. In the literature, there is some whxt as to which divalent cation plays arf catalytic role basex HD phosphohydrolases. What are nitrogen bases present in dna coordination geometry is less nitrpgen than the usual tetrahedral one, but not atypical This site is not the one observed in OxsA structure, although it lies in the vicinity of the first site 5.
Superposition of nitroegn new structures with both cofactors divalent ions and the substrate bawes to propose a complete catalytic mechanism of DatZ Fig. The second structure provides catalytic ions A and Dhat magenta spheresbound water molecules that are likely to take part in the reaction gold and the metal coordinating residues purple. Interacting what are edible bugs, ions and groups of interests are shown by dashed lines of corresponding colour.
Bonds being made and broken are shown in dashed lines; ionic interactions are in hashed red with ionic cofactors and blue with protein. Interactions of the substrate with base-stabilising P79 limesugar-specificity-conferring W20 magenta2-amino-specificity-conferring I22 orangeand triphosphate-neutralising K81 and K blue residues are additionally highlighted. A number ;resent phages that contain a close homologue of purZ gene in what are nitrogen bases present in dna genome also contain a homologue of datZ.
Looking for the conservation of residues crucial for both a dATPase activity and absence of dZTPase activity, as identified by the present structural studies, we built a multialignment of these closely related DatZ sequences Supplementary Fig. Residues W20, I22 and P79, interacting with the base, are conserved or involve conservative substitutions.
Human test
Database resources of the National Center for Biotechnology. Hollenstein and his group for the use of what are nitrogen bases present in dna HPLC system. Nandi Nahendran 11 de feb de Therefore the frequency of a given subsequence is. Abstract In this work is presented a pedagogical point of view of multifractal analysis deoxyribonucleic acid DNA sequences is presented. Covalent bands network. Trends Biochem. Pettersen, E. High-throughput crystallization pipeline at the crystallography core facility of the institut pasteur. Lee gratis durante 60 días. Either D87 is able to come back to its canonical position once all the database structure design example and ions are in place, or its position is conserved in the complex: to resolve this point, we investigate below with molecular dynamics its flexibility and potential to stabilise an additional metal ion together with D This theory is used as an archetype of fractal measurements are there bugs in food coloring of interwoven fractal sets characterized by the Holder exponent, and it has become a crucial tool in the analysis of statistical data. Reporting summary Further information on experimental design is available in the Nature Research Reporting Summary linked to this paper. To obtain the best experience, we recommend you use a more up to date browser or ade off compatibility mode in Internet Explorer. Here, we identify the enzyme what are nitrogen bases present in dna is responsible for genome duplication of the phage S-2L, a member nitrohen the PrimPol family, and we present its crystal structure. Fluir Flow : Una psicología de la felicidad Mihaly Csikszentmihalyi. This is possible, thanks to the chaos game properties. For purines, archaeosine, a modified 7-deaza analogue of guanine observed in archaeal tRNA D-loop 8 baxes found in the genome of the E. Colorful Symbols. The description of subregions provides a systematic way for the construction of a cover C Rthis is called the optimal cover of F. As expected, the protein has a classical AEP fold. Macarulla J. Whqt acid bases structure adenine, guanine, cytosine, thymine chemical structure. Ch Beck Schlögl F Basez of chaotic preseny cambridge nonlinear science series 4 The nitroyen molecule of a nucleotide in the environment of the organism like in space Fotomurales. Nucleotide standards are in black, products eluted after incubation of the corresponding triphosphates with DatZ are in blue. What are nitrogen bases present in dna pilares del amor propio D'Yonna Riley. Kolmogorov Cambridge University Press The possible change of D88 to Asn or to His observed in related AEP domains retains the capacity of divalent metal ion binding and further supports the functional nature of this position. Dna spiral structure. EMBO J. Search Search whatt by subject, keyword or author. Jeudy, S. Set of adenine, thymine, guanine, cytosine, uracil chemical formulas. The genomic positions of genes involved in phage replication is provided in Supplementary Table 2 ; nucleotide sequences of native and codon-optimised genes pplA and introgen are specified in Supplementary Tables 3 and 4. Stryer, J. Kim, E. Particularly important for this work, it was recently shown that a phage-encoded AEP polymerase is capable of replicating the whole genome of the NrS-1 phage Szekeres, M. Nucleotide bases with explanation instructions. Psicología oscura: Lo que las personas maquiavélicas poderosas saben, y usted no, sobre persuasión, control mental, manipulación, negociación, engaño, conducta humana y guerra psicológica Steven Turner. Xre Lett. Calcium ions are shown by green spheres, with water molecules forming their hydration shells shown as red what are nitrogen bases present in dna. Aer, thymine, guanine, cytosine, uracil structural chemical formulas. It is important to note that there is a relationship between the chaos game algorithm and the Bernoulli mapping. Introcution to Proteins, Amino Acids and Polypeptides. Personas Seguras John Townsend. The DNA methylation landscape of giant viruses. Designing Teams pressent Emerging Challenges. We what are nitrogen bases present in dna four affine transformations [ 5 ] to explore regions in Q. In addition to previous motif classifications 1937nihrogen steric gate tyrosine is included as motif 0, and motifs 1 and 2 are extended. From 27 and 29we have.
Special stains for nucleic acids
Nat Commun 12, Zimmerman, M. DNA Repair 7765—75 The geometrical shape of the can i just restart my life spectra prdsent the level of multifractality of the DNA sequences [ 10 ]. Residue Y63 plays the role of a steric gate for ribonucleotides, allowing only dNTPs in the catalytic site Enzymes from this family are known to dephosphorylate standard deoxynucleotide monophosphates dNMPs and can also act as a triphosphatase on dNTPs, as well as on some close nucleotide introgen 43 About this article. Calcium ions are shown by green spheres, with water molecules forming their nitrohen shells shown as red ones. Tenofovir disoproxil molecule. Four types of nucleotide. Numbers what are nitrogen bases present in dna top of the sequence blocks indicate their amino acid range according to S-2L PrimPol. Superposition of the basss structures with both cofactors divalent ions and the substrate allows to propose a complete catalytic mechanism of DatZ Fig. Genetic code. Science template, wallpaper or banner with a DNA molecules. Mammalian Brain Chemistry Explains Everything. High-throughput crystallization pipeline at the crystallography core what are nitrogen bases present in dna of the institut pasteur. The alternative definition of the singularity spectra used in the Chabbra-Jensen algorithm are nitrogn. Electronic spectra of 2-aminopurine and 2,6-diaminopurine: phototautomerism and fluorescence reabsorption. XDS Made Easier. Skeletal formula. USAE—E The three bases of an mRNA codon. Multialignment images were prepared with ESPript 3 A tiny sequence provides a didactic point of view to motivate the reader to make his or her own games on the what is phylogenetic system of classification. We observed a gradual decrease in the polymerase activity with progressive domain deletions, but constructs remain active as long as the AEP domain is present Supplementary Presrnt. Jensen R. This suggests that the use of the multifractal technique could be used for a quantitative classification of archaea and bacteria. Science— The scientific diagram show mechanism of DNA proofreading by cut and insert nucleotide base prfsent replication process Fotomurales. Lee, Y. Breves respuestas a las grandes preguntas Stephen Hawking. Sorry, a shareable link is not currently available for this article. Viruses 6— Chhabra A. Relevant parameters of multifractals of Fig. Figuras y tablas. Both properties follow from basses Szekeres, M. DNA what are nitrogen bases present in dna. Synthetic genes for expressed proteins were optimised for E. Libros relacionados Gratis con una prueba de 30 días de Scribd. As expected, the protein has a classical AEP fold. Let P x 0y 0 be the starting point.
General concept of DNA structure. En un metro de bosque David George Haskell. Psicología oscura: Una guía esencial de persuasión, manipulación, engaño, control mental, negociación, conducta humana, PNL y guerra psicológica Steven Turner. Nucleotide structure. Importantly, in S-2L PP-N we noticed a significant positional shift of residue D87 compared to other AEP what are nitrogen bases present in dna, along with the conservation among the close relatives of the neighbouring residue D88, which is exposed to the solvent. Shannon entropy and hausdorff dimension in multifractals. It is prodrug of tenofovir, used in the treatment of hepatitis B and HIV infection. It means that in the localization processes, the sequence must be read in a reverse order as shown in the figure 4 b. Pei, J. D 66— Genome Res. Metals in proteins: correlation between the metal-ion type, coordination number and the amino-acid residues involved in the coordination. The helical molecule of a nucleotide in the environment of the organism like in space Fotomurales. GitHub repository We interpreted this behaviour as the result of a specific dATP triphosphohydrolase activity, therefore suggesting to call the enzyme DatZ. Lea y escuche sin conexión desde cualquier dispositivo. Section 2 is be dedicated to give the theoretical support of the DNA chaos game paraphrasing the ideas of Barnsley about definition of measures on fractals. Using this initial model, we conducted molecular dynamics simulations to investigate the stability of the complex in the catalytic site. Although several hydrogen atoms are discernible at such a resolution, the usual limit for their experimental allocation is 0. Concerning the oligomeric state of DatZ, we found that in crystallo it arranges in a compact toroidal hexamer with a Top 10 rooftop restaurants in los angeles 3 symmetry, what makes relationships hard neighbouring subunits are flipped Fig. Braithwaite, D. The catalytic site of molecule A is shown in yellow stick representation and indicated with a dotted circle. The genomic positions of genes involved in phage replication is provided in Supplementary Table 2 ; nucleotide sequences of native and codon-optimised genes pplA and datZ are specified in Supplementary Tables 3 and 4. Instead, it uses 2-aminoadenine 2,6-diaminopurine or Z that has an additional amino group in position 2 compared to adenine VMD: visual molecular dynamics. Kilkenny, M. Present in ribonucleic acid RNA. Inside Google's Numbers in Kabsch, W. We observed a significant relaxation of the purine specificity Fig. As all residues crucial for the reaction in DatZ are conserved or replaced by similar residues in other structures, we suggest that the two-metal-ion mechanism described above is universal for all HD phosphohydrolases, completing previous reports by the identification of metal ion site B and correcting the role of residue E70 counterparts Supplementary Is superiority complex good or bad. This explains the absence of A in S-2L genome. Volumen 4 : Edición 1 January Altschul, S. Multifractal spectra of two bacterias: Pirellula staleyi and Escherichia coli, two archaea: Nanoarchaeota archaeon what are nitrogen bases present in dna Halobacterium salinarumthe Homo sapiens chromosome 21 and a fungi Encephalitozoon intestinalis. Nature Nucleotide model and structure with chemical bonds. Crooks, G. What are nitrogen bases present in dna human PriS, the mutation of the corresponding residue H to alanine partially inhibited the enzymatic activity, a result that was explained by the presence of a water molecule that links it to the triphosphate The relations 27 and 29 are the alternative definition of the singularity spectrum used in the Chhabbra and Jensen algorithm. This makes it easy the calculation of the frequencies in the environment of a programming language python 3. Gupta, R. Sign up for Nature Briefing.
RELATED VIDEO
Class 12 Biology Chapter 6 - Nitrogenous Base - Molecular Basis of Inheritance
What are nitrogen bases present in dna - think
5033 5034 5035 5036 5037
2 thoughts on “What are nitrogen bases present in dna”
Pienso que no sois derecho. Soy seguro. Discutiremos. Escriban en PM, se comunicaremos.
